| dyads_test_vs_ctrl_m1_shift8 (dyads_test_vs_ctrl_m1) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
; dyads_test_vs_ctrl_m1; m=0 (reference); ncol1=14; shift=8; ncol=23; --------wtcACACGTGTgaw-
; Alignment reference
a 0 0 0 0 0 0 0 0 158 112 78 383 27 452 12 34 16 23 33 100 163 128 0
c 0 0 0 0 0 0 0 0 109 124 217 59 434 7 441 9 24 14 26 103 101 105 0
g 0 0 0 0 0 0 0 0 105 99 103 23 14 24 9 443 7 435 60 220 122 111 0
t 0 0 0 0 0 0 0 0 126 163 100 33 23 15 36 12 451 26 379 75 112 154 0
|
| BAM8.DAP_M0154_AthalianaCistrome_shift9 (BAM8.DAP:M0154:AthalianaCistrome) |
 |
0.930 |
0.755 |
6.951 |
0.950 |
0.968 |
8 |
4 |
7 |
7 |
9 |
7.000 |
1 |
; dyads_test_vs_ctrl_m1 versus BAM8.DAP_M0154_AthalianaCistrome (BAM8.DAP:M0154:AthalianaCistrome); m=1/109; ncol2=15; w=13; offset=1; strand=D; shift=9; score= 7; ---------yCACACGTGysaam
; cor=0.930; Ncor=0.755; logoDP=6.951; NsEucl=0.950; NSW=0.968; rcor=8; rNcor=4; rlogoDP=7; rNsEucl=7; rNSW=9; rank_mean=7.000; match_rank=1
a 0 0 0 0 0 0 0 0 0 34 19 417 0 599 2 0 0 13 29 84 295 269 167
c 0 0 0 0 0 0 0 0 0 206 499 37 599 0 598 0 10 0 244 241 73 99 195
g 0 0 0 0 0 0 0 0 0 5 42 139 0 1 0 600 0 585 12 248 93 123 100
t 0 0 0 0 0 0 0 0 0 355 40 7 1 0 0 0 590 2 315 27 139 109 138
|
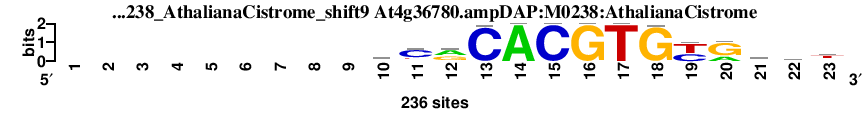
| At4g36780.ampDAP_M0238_AthalianaCistrome_shift9 (At4g36780.ampDAP:M0238:AthalianaCistrome) |
 |
0.919 |
0.796 |
7.053 |
0.948 |
0.965 |
13 |
2 |
4 |
10 |
15 |
8.800 |
2 |
; dyads_test_vs_ctrl_m1 versus At4g36780.ampDAP_M0238_AthalianaCistrome (At4g36780.ampDAP:M0238:AthalianaCistrome); m=2/109; ncol2=14; w=13; offset=1; strand=D; shift=9; score= 8.8; ---------tCrCACGTGyRsat
; cor=0.919; Ncor=0.796; logoDP=7.053; NsEucl=0.948; NSW=0.965; rcor=13; rNcor=2; rlogoDP=4; rNsEucl=10; rNSW=15; rank_mean=8.800; match_rank=2
a 0 0 0 0 0 0 0 0 0 54 10 121 0 236 0 0 0 0 0 67 36 91 23
c 0 0 0 0 0 0 0 0 0 50 162 12 232 0 236 0 0 2 105 0 98 58 39
g 0 0 0 0 0 0 0 0 0 26 26 98 0 0 0 236 0 233 0 166 75 46 39
t 0 0 0 0 0 0 0 0 0 106 38 5 4 0 0 0 236 1 131 3 27 41 135
|
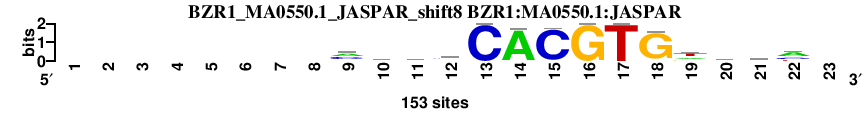
| BZR1_MA0550.1_JASPAR_shift8 (BZR1:MA0550.1:JASPAR) |
 |
0.883 |
0.883 |
6.304 |
0.948 |
0.962 |
34 |
1 |
33 |
13 |
21 |
20.400 |
10 |
; dyads_test_vs_ctrl_m1 versus BZR1_MA0550.1_JASPAR (BZR1:MA0550.1:JASPAR); m=10/109; ncol2=14; w=14; offset=0; strand=D; shift=8; score= 20.4; --------mmkmCACGTGdvmm-
; cor=0.883; Ncor=0.883; logoDP=6.304; NsEucl=0.948; NSW=0.962; rcor=34; rNcor=1; rlogoDP=33; rNsEucl=13; rNSW=21; rank_mean=20.400; match_rank=10
a 0 0 0 0 0 0 0 0 63 55 30 49 0 147 6 0 0 6 42 41 42 81 0
c 0 0 0 0 0 0 0 0 60 47 20 66 153 2 147 0 0 5 0 53 61 39 0
g 0 0 0 0 0 0 0 0 30 27 52 26 0 0 0 153 0 142 39 43 22 0 0
t 0 0 0 0 0 0 0 0 0 24 51 12 0 4 0 0 153 0 72 16 28 33 0
|
| BAM8.ampDAP_M0155_AthalianaCistrome_rc_shift9 (BAM8.ampDAP:M0155:AthalianaCistrome_rc) |
 |
0.936 |
0.761 |
0.290 |
0.953 |
0.972 |
6 |
3 |
103 |
5 |
7 |
24.800 |
15 |
; dyads_test_vs_ctrl_m1 versus BAM8.ampDAP_M0155_AthalianaCistrome_rc (BAM8.ampDAP:M0155:AthalianaCistrome_rc); m=15/109; ncol2=15; w=13; offset=1; strand=R; shift=9; score= 24.8; ---------ysACACGTGysaay
; cor=0.936; Ncor=0.761; logoDP=0.290; NsEucl=0.953; NSW=0.972; rcor=6; rNcor=3; rlogoDP=103; rNsEucl=5; rNSW=7; rank_mean=24.800; match_rank=15
a 0 0 0 0 0 0 0 0 0 78 13 428 0 588 0 0 0 26 5 104 305 280 141
c 0 0 0 0 0 0 0 0 0 188 397 44 590 0 590 0 0 0 289 174 72 114 166
g 0 0 0 0 0 0 0 0 0 7 161 81 0 2 0 590 0 564 0 268 87 129 132
t 0 0 0 0 0 0 0 0 0 317 19 37 0 0 0 0 590 0 296 44 126 67 151
|




